- Definition
- Viruses are obligate intracellular parasites, meaning they require a host cell’s machinery to replicate.
- A complete infectious virus particle is called a virion.
- Structure & Classification
- Genome: Can be DNA or RNA, which can be double-stranded (ds) or single-stranded (ss), and linear or circular. All viruses are haploid (one copy of the genome), except for retroviruses, which are diploid.
- Capsid: A protein coat that encloses the viral genome. The capsid plus the genome is the nucleocapsid.
- Icosahedral: A complex, spherical-appearing structure.
- Helical: A spiral arrangement of proteins.
- Envelope: A lipid bilayer derived from the host cell membrane that surrounds the capsid in some viruses (enveloped viruses). Those without it are called naked viruses.
- Enveloped viruses are sensitive to alcohol, detergents, and ether.
- Enveloped Viruses = Budding/Exocytosis (Host Cell Usually Survives)
- Naked Viruses = Lysis (Host Cell Destruction)
- General Rules & Exceptions:
- All DNA viruses are dsDNA, except for Parvoviridae (ssDNA).
- All DNA viruses replicate in the nucleus, except for Poxviridae (replicates in the cytoplasm).
- They must use the host’s DNA and RNA polymerases, which are in the nucleus.
- Poxvirus: It’s a massive, complex virus. It’s so large that it encodes its own DNA-dependent RNA polymerase and other replication machinery. It’s self-sufficient and doesn’t need to enter the nucleus.
- All RNA viruses are ssRNA, except for Reoviridae (dsRNA).
- Most RNA viruses replicate in the cytoplasm, except for Orthomyxoviridae (Influenza virus) and Retroviridae, which replicate in the nucleus.
- They bring their own enzyme (RNA-dependent RNA polymerase, host doesn’t have) and only need host ribosomes, which are in the cytoplasm.
- Influenza: It has a segmented genome and uses a unique mechanism called “cap-snatching,” where it steals the 5’ caps from host pre-mRNAs in the nucleus to use as primers for its own mRNA synthesis.
- Viral Replication Cycle
- Attachment: Virus binds to specific receptors on the host cell surface, which determines viral tropism (the type of cells a virus can infect).
- Penetration: The virus enters the host cell, often through endocytosis or fusion of the viral envelope with the cell membrane.
- Uncoating: The viral capsid is degraded, releasing the nucleic acid into the host cell.
- Synthesis: The virus hijacks host machinery to replicate its genome and synthesize viral proteins.
- Positive-sense (+ssRNA) viruses: Their genome can be directly translated by host ribosomes, similar to mRNA.
- Negative-sense (-ssRNA) viruses: Must carry their own RNA-dependent RNA polymerase to transcribe their genome into a positive-sense strand before protein synthesis can occur.
- Retroviruses (e.g., HIV): Use a viral reverse transcriptase (an RNA-dependent DNA polymerase) to convert their RNA genome into DNA, which is then integrated into the host genome.
- Assembly: New viral components are assembled into progeny virions.
- Release: New viruses exit the host cell.
- Naked viruses are typically released through cell lysis.
- Enveloped viruses are released via budding, acquiring their envelope from the host cell membrane.
- Viral Genetics
- Recombination: Exchange of genetic material between two homologous viral genomes.
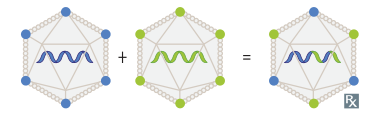
- Reassortment: The mixing of genome segments when a host cell is co-infected with two segmented viruses (e.g., Influenza virus, Rotavirus). This can lead to sudden, major changes known as antigenic shift, which can cause pandemics. t
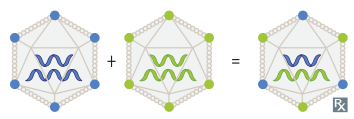
- Genetic Drift: Minor changes due to point mutations in the viral genome over time (e.g., seen in influenza).
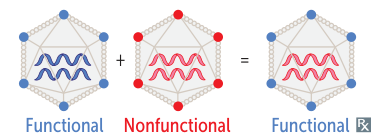
- Complementation: When one of two viruses infecting a cell has a mutation that results in a non-functional protein, the other virus can “complement” it by making a functional version of that protein, allowing both viruses to replicate.
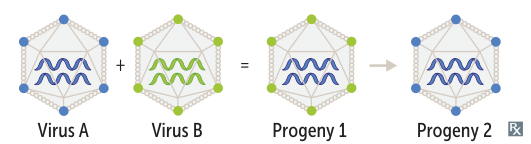
- Phenotypic Mixing: Progeny virions contain the genome of one virus but are coated with surface proteins from another virus that co-infected the cell. The next generation of viruses will revert to their original phenotype, as the genome is unchanged.
- Laboratory Diagnosis
- Nucleic Acid Amplification Tests (NAATs): Techniques like PCR are highly sensitive and specific for detecting viral DNA or RNA. “Viral load” quantification is crucial for managing infections like HIV and HCV.
- Antigen Detection: Direct detection of viral proteins in clinical samples using methods like ELISA or immunofluorescence. Faster but less sensitive than NAATs.
- Serology: Detects host antibodies (IgM or IgG) against the virus. An IgM response suggests a recent or acute infection, while IgG indicates a past infection or vaccination.
- Viral Culture: Growing the virus in cell lines, which can cause visible changes known as cytopathic effects (CPE). Historically the gold standard but slow.



