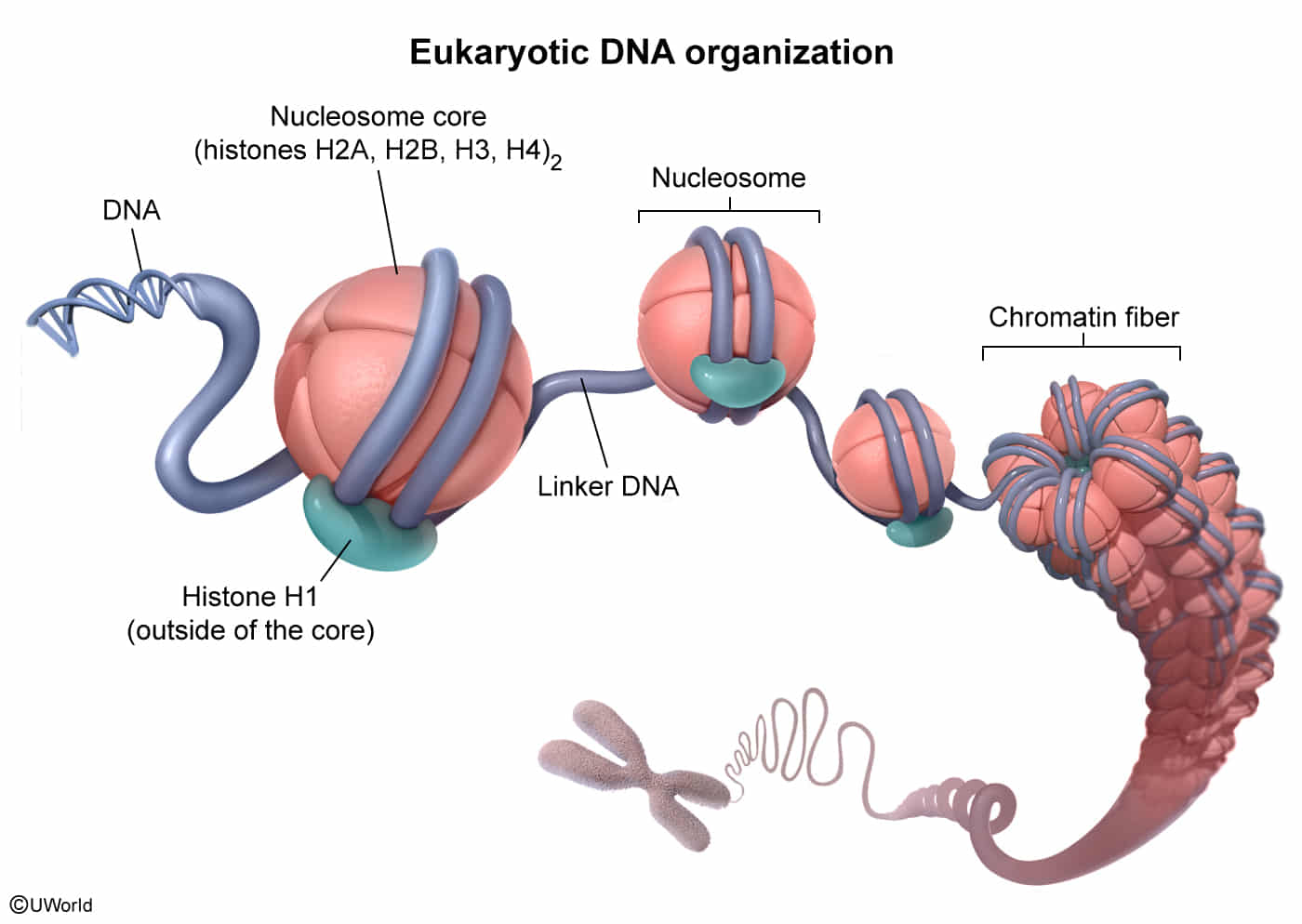
- Structure & Composition
- Small, basic proteins rich in Lysine and Arginine (positively charged).
- Bind to the negatively charged phosphate backbone of DNA.
- Nucleosome Core: Octamer consisting of (H2A, H2B, H3, H4) x 2. DNA wraps around this core ~1.75 times.
- Histone H1 (Linker): Binds to the nucleosome and “linker DNA” to stabilize the chromatin fiber. It is outside the core octamer. t
- Epigenetic Regulation
- Acetylation
- Performed by Histone Acetyltransferases (HATs).
- Removes (+) charge from lysine → relaxes DNA coiling → Euchromatin.
- Result: ↑ Transcription (“Histone Acetylation makes DNA Active”).
- Deacetylation
- Performed by Histone Deacetylases (HDACs).
- Restores (+) charge → tightens DNA coiling → Heterochromatin.
- Result: ↓ Transcription (Gene silencing).
- Methylation
- Histone methylation can repress or activate transcription depending on the specific location.
- Contrast with DNA Methylation (CpG islands): “Methylation makes DNA Mute” (silencing/imprinting).
- Clinical Relevance
- Drug-Induced Lupus Erythematosus (DILE)
- Classic triggers: Hydralazine, Procainamide, Isoniazid.
- Often seen in Slow Acetylators.
- Serology: Anti-histone antibodies are characteristic (>95% sensitive).
- Huntington Disease
- Mechanism involves Histone Deacetylation → silencing of genes necessary for neuronal survival → neuronal cell death.
- Mitochondrial DNA
- Unlike nuclear DNA, mitochondrial DNA is circular and does not utilize histones.
